Molecular evolution
Recall that teh gene tree need not match the species tree.
One case in which this is likely is when alternate alleles have persisted in two species since the speciation event.

Note that here, some alleles in species A are more closely related to some alleles in species B than to other alleles at the same locus in species A. Thus, at that locus, each species is "polyphyletic".
Example: HLA genes (Human Leukocyte Antigen)
These are involved with self recognition in the immune system.
There are many alleles with heterozygote superiority.
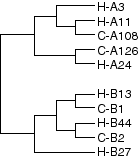
Humans have two loci, HLA-A and HLA-B
Chimps have two homologous loci, C-A and C-B
This suggests that there has been strong balancing selection at the HLA loci for the past 6-7 million years.
Constructing a phylogeny based on sequence data produces this tree for Human (H) and Chimp (C) alleles.
Identifying selection on genes
Recall that some base pair changes in a DNA sequence do not lead to any
amino acid change in a resulting protein; such changes are called
"synonymous" changes.
By contrast, "non synonymous" DNA substitutions are those that do
lead to an amino acid change.
Define:
(Note: You will sometimes see the terms "dn" and "ds" instead of KA and KS. These refer to the rates (number of changes per unit time interval) rather than the actual number of changes. However, since the time since the common ancestor is the same for both species, considering dn/ds yields the same result as KA/KS.)
If a gene is completely neutral, so that all possible mutations of it have no effect on fitness, then we expect that KA/KS = 1.
If KA/KS < 1, then non synonymous substitutions are less likely to be fixed than are synonymous substitutions. This suggests that "purifying selection" has been acting on the gene, meaning that most mutations that lead to an amino acid change are selected against.
In KA/KS > 1, then mutations that change the amino acid sequence are being fixed at a higher rate than neutral mutations. This suggests directional selection acting on the gene.
Detecting alleles that are currently spreading through a population due to selection
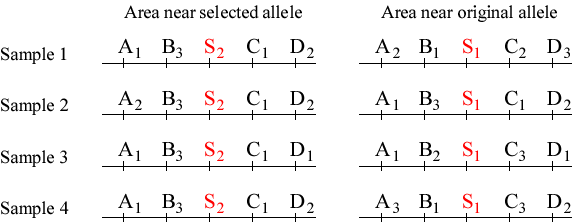
If an allele spreads rapidly through a population, it carries with it linked regions of the genome (these are said to be hitchhiking).
A consequence of this is that if we look in the chromosomal neighborhood of an
allele that is rapidly increasing in frequency, we see much less variation in
nearby loci than in more distant parts of the genome.
By contrast, if we look at a homologous chromosome that carries the older
allele (that is now declining in frequency), we see much more variation at nearby
loci.
A hypothetical example: Allele S2 is rapidly increasing in frequency due to selection, and is replacing allele S1. We consider the pattern of variation at sites A, B, C, and D, that surround the S locus.
This approach to detecting genes under active selection has recently been
applied to the human genome.
This has identified a number of genes that appear to actively evolving in
different populations.
These include genes that influence traits like bone structure, skin color, ability to digest lactose, fertility, and some brain functions.
Notably, different populations tend to show different genes being selected.
New Genes
So far, we have considered mutations of existing genes. Now we consider
where new genes themselves come from.
Gene Duplication
This can occur through unequal crossing over.
If homologous chromosomes are offset when they pair in Meiosis, then
a crossover event can produce one chromosome with a duplicated gene and
another where the gene is lost.
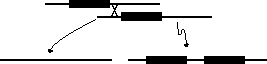
If the gene performs an adaptive function, then the chromosome that has lost it will itself be lost. The chromosome with two copies, however, may persist.
Genes related by such a duplication event are called "Paralogous".
Genes in separate species that have a common ancestor are "Orthologous".
Possible outcomes:
Divergence may occur after, during, or before duplication.
Divergence after duplication:
Usually this will produce a pseudogene.
However, if a variant of the existing protein is adaptive in concert
with the initial form, then mutations on one or the other copy can produce
a new functional gene.
Divergence during duplication:
Sometimes, gene expression is influenced by position on the chromosome.
In such a case, the mere fact that the duplicate gene is in a different
location may cause it to be expressed differently - either to a greater
or lesser degree or at a different time in development (see Hox genes discussed
later).
Divergence before duplication:
If there is heterozygote superiority, then it is adaptive to have two
different variants of a protein in the same individual.
This can become permanent if an unequal crossover puts two different
alleles on the same chromosome.
Some examples clearly show evidence of gene duplication.
Example 1: Globin gene families.
Myoglobin
On Chromosome 22. Stores O2 in muscles.
Hemoglobin (alpha and beta)
Form a tetramer (2 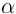 and 2
and 2 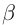 chains)
that transports O2.
Jul 8, 2021
chains)
that transports O2.
Jul 8, 2021