Form a tetramer (2
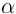 and 2
and 2 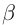 chains)
that transports O2.
chains)
that transports O2.
Recall the Globin gene families.
Myoglobin
On Chromosome 22. Stores O2 in muscles.
Hemoglobin (alpha and beta)
Form a tetramer (2 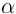 and 2
and 2 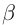 chains)
that transports O2.
chains)
that transports O2.
Molecular clock indicates that these are all derived from a common ancestral gene 600-800mya (take this with a large grain of salt)
In most vertebrates, functional hemoglobin is composed of four different
molecules, 2 alpha and 2 beta hemoglobins.
These diverged 450-500 mya.
This supported both by molecular clock and the fact
that Jawless Fish have only alpha hemoglobin.
Both alpha and beta are really multigene families, with many copies.
Within each family, some copies are expressed at different stages in
development.
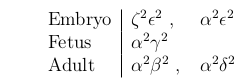
Duplication has allowed diversification of function.
For example, 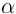 2
2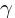 2
(Fetal) hemoglobin has a higher oxygen binding affinity than adult hemoglobin.
2
(Fetal) hemoglobin has a higher oxygen binding affinity than adult hemoglobin.
This is adaptive since the fetus must extract oxygen from the
mother's circulatory system.
(The higher O2 binding affinity of 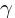 hemoglobin is the result of a single AA substitution, a serine instead of a
histidine at position 143).
hemoglobin is the result of a single AA substitution, a serine instead of a
histidine at position 143).
The phylogeny of some of the globin genes found in humans looks like this:
Note:
1) Modern Jawless fish, which branched off around 500 mya (based on fossil evidence) have only alpha (no beta) hemoglobin.
2) The alpha chain diversified about the time that vertebrates came on land.
3) The gamma chain branched off from the beta line about the time that placental mammals arose.
4) Molecular clock estimates put the divergence of the two types of gamma globin at about 30 million years ago. This is consistent with the fact that while humans and old world monkeys have both types, new world monkeys have only one type of gamma globin.
Example 2: Color sensitive pigments in primates.
Apes (including humans) and old world monkeys have three pigments:
New World Monkeys (which diverged before the split between Old World
Monkeys and Apes) have an autosomal Blue pigment gene but only one X linked
pigment gene (generally Red).
Thus, most can not distinguish as many colors as
we can.
But...
In Squirrel Monkeys (New World) there are two different alleles of
the X linked gene, one Red and one Green.
Females that are heterozygous at this locus thus have full trichromatic
color vision.
Exon Shuffling
In addition to duplication of genes, new genes can emerge through the reshuffling of parts of existing genes.
Recall the basic structure of a eucaryotic gene:
5' end - Promoter region - Transcribed region - Stop codon - Flanking region
- 3' end
The transcribed region is composed of:
Exons, which are generally translated to a protein sequence.
Introns, which are excised from the pre-mRNA.
Exons sometimes correspond to functional domains in proteins.
Since exons are separated by excised introns, they can sometimes be
shuffled around, potentially producing new protein functions.
Rearranging functional domains of proteins is much more likely to produce
an adaptive variant than random changes within the coding regions.
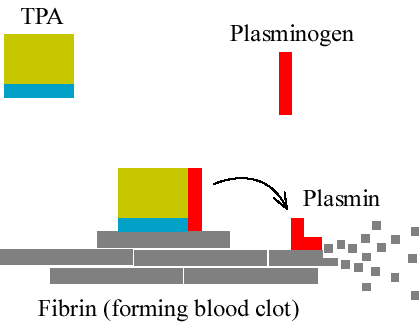
Example: Tissue Plasminogen Activator (tPA)
TPA converts Plasminogen to Plasmin.
Plasmin dissolves Fibrin, a protein in blood clots.
The action of tPA is accelerated by Fibrin itself, so TPA activity is enhanced when there is need for it.
This happens because Fibrin binds to both tPA and Plasminogen, bringing them together.
A related Plasminogen activator, Urokinase (UK), does not bind Fibrin.
There are many copies of the UK gene.
TPA and UK have similar sequences except for a 43 Amino Acid
sequence found in tPA but not UK.
This sequence is the Fibrin binding region.
This same 43 AA sequence is found in another protein, Fibronectin, that binds Fibrin for other reasons.
In both tPA and Fibronectin, the Fibrin binding region corresponds to an exon, that appears to have been transferred from one gene to the other.
Transposable Elements
Coding genes account for <10% of the DNA in many eucaryotic genomes.
Some of the non-coding DNA is in the form of pseudogenes (discussed earlier) and repeated sequences.
Much is in the form of transposable elements
Around 45% of the human genome is made up of transposable elements.
Types:
Reverse transcriptase:
Makes a complimentary DNA strand (cDNA) from RNA.
Retroposons make an RNA transcript which is then reverse transcribed
back to DNA and inserts elsewhere in the genome.
Retroposons include LINEs (Long Interspersed Elements) and SINEs (Short Interspersed Elements).
Example: Movement of exons by LINES.
LINE-1 elements comprise ~15% of the human genome.
The sequence that terminates transcription in LINES (the poly-A tail) is "weak" in the sense that transcription sometimes proceeds through it.
In such cases, downstream sequences are transcribed with the LINE element until a more substantial poly-A region is encountered.
This extra RNA can contain exons, which are then resinserted into the DNA at some other location by reverse transcription. Jul 8, 2021