Innovative Research

Texas Tech University biologist Caleb Phillips.
Caleb Phillips Finds Genetics May Rule Wound Infections
Biologist Says Genes of Pathogens in Chronic Wounds Can Hinder the Healing Process.
6.24.2020 by Amanda Castro-Crist
Think of the last time you had a cut on your leg or foot. How long did it take for that wound to show signs of healing? If it was longer than three weeks, doctors would refer to it as a "chronic wound." These types of non-healing wounds affect millions of people and are often associated with other chronic diseases or conditions like diabetes, decreased circulation and neuropathy.
But now, in a first-of-its-kind study, researchers have determined that genetics may play a role in how wounds heal. Caleb Phillips, an assistant professor at Texas Tech University and director of the Phillips Laboratory in the Department of Biological Sciences, and doctoral student Craig Tipton led the study, "Patient genetics is linked to chronic wound microbiome composition and healing," published June 18, 2020, in the open-access, peer-reviewed medical journal PLOS Pathogens.
Phillips, who also serves as the Curator of Genetic Resources at the Natural Science Research Laboratory's (NSRL) Robert J. Baker Genetic Resources Collection, said the study determined that certain genes are associated with the number of bacteria and abundance of common pathogens in wounds. The collection of microbes, known as a "microbiome," can determine how a wound heals and how long that process takes. The research also showed that the more diversity within a wound microbiome, the less time it took to heal.
"A chronic wound is a serious burden," Phillips said. "The median healing time of patients in this study was more than 200 days, but some people deal with these wounds for years. We were able to show that a person's genetics explain differences in the species that infect their wounds. The information in this study could be valuable in a clinical setting as pre-operative information to help inform preventative measures before a procedure, as some chronic wounds arise as non-healing surgical wounds, and could help inform a course of treatment for an existing infection."
Researchers included colleagues from Texas Tech, the Texas Tech University Health Sciences Center (TTUHSC), Lubbock's Southwest Regional Wound Care Center (SWRWCC) and the University of North Texas Health Science Center at Fort Worth (UNTHSC). Phillips said the project began after a conversation with Dr. Randy Wolcott, founder of the SWRWCC, where they discussed how certain patients develop multiple non-healing wounds over a long period. The microbiome within each wound was essentially the same. The researchers wanted to find out if this could be partially explained by genetics.
"This study is big," Wolcott said. "It's the initial study to find genes and/or gene alterations that correlate with what bacterial species can be more successful in causing infections in a specific patient. If a screen of a patient's DNA prior to a surgery showed that patient is highly susceptible to a staphylococcus species, the doctor could mitigate staphylococcus complications."
Patients visiting the SWRWCC for the care of a lower-extremity infected wound consented to participation in the study and provided samples from their wound(s) and from a cheek swab. Samples like those collected at the SWRWCC are archived in liquid nitrogen at the Wolcott Wound Care Research Collection, a collection of the NSRL's Genetic Resources Collection that is specifically dedicated to wound care biology. The design of the study included an exploratory cohort of 79 patients, from which "candidate locations" of their genome were identified. This was followed by an experimental cohort of 85 patients, used to confirm associations between the genomic locations and wound microbiome characteristics.
The bacterial communities infecting an individual's wounds were determined by microbiome sequencing methods, and each patient's genome was characterized at a few hundred thousand specific locations, called "single nucleotide polymorphisms," or SNPs. A statistical approach was then used to determine which of these genomic locations explained differences in an individual's wound microbiome composition and was followed with several downstream analyses to understand the results.
"We showed that there are identifiable locations in people's genome where, depending on their genotype, they tend to get infections by specific bacteria," Phillips said. "The different genomic locations identified tend to be related in terms of the types of genes they are close to and may regulate. A working hypothesis emerging from the research is that genetic differences influencing genes encoding the way our cells interact with the environment and each other are important for infection differences."
 Tipton, who completed his bachelor's degree in biology at Angelo State University
before arriving at Texas Tech, said the project has been a significant part of his
dissertation, which focuses on learning more about why a person's wounds are infected
by different types of microbes. Though there is still work to be done before the research
directly benefits patients, Tipton said the study is an important and promising step
in that direction.
Tipton, who completed his bachelor's degree in biology at Angelo State University
before arriving at Texas Tech, said the project has been a significant part of his
dissertation, which focuses on learning more about why a person's wounds are infected
by different types of microbes. Though there is still work to be done before the research
directly benefits patients, Tipton said the study is an important and promising step
in that direction.
"Personalized medicine is a current hot topic in modern healthcare, where the goal is to identify inherent differences within individuals that may cause them to be impacted differently by disease and finding treatments that are well-suited and tailored to the individual and may contribute to better patient outcomes," Tipton said. "Our project furthers two equally-interesting avenues of research with potential translation to the clinic. In one, it is our goal to develop robust genomic predictive models that could help physicians to determine a patient's risk for chronic wound infection, particularly to specific bacteria.
"In the second, this work helps to inform how genetic variation in patients can influence microbiome-host interactions and wound infection pathogenesis. By further studying infection pathogenesis and how these complex microbial communities interact, it may be possible to improve existing therapies or to develop new therapeutic strategies altogether."
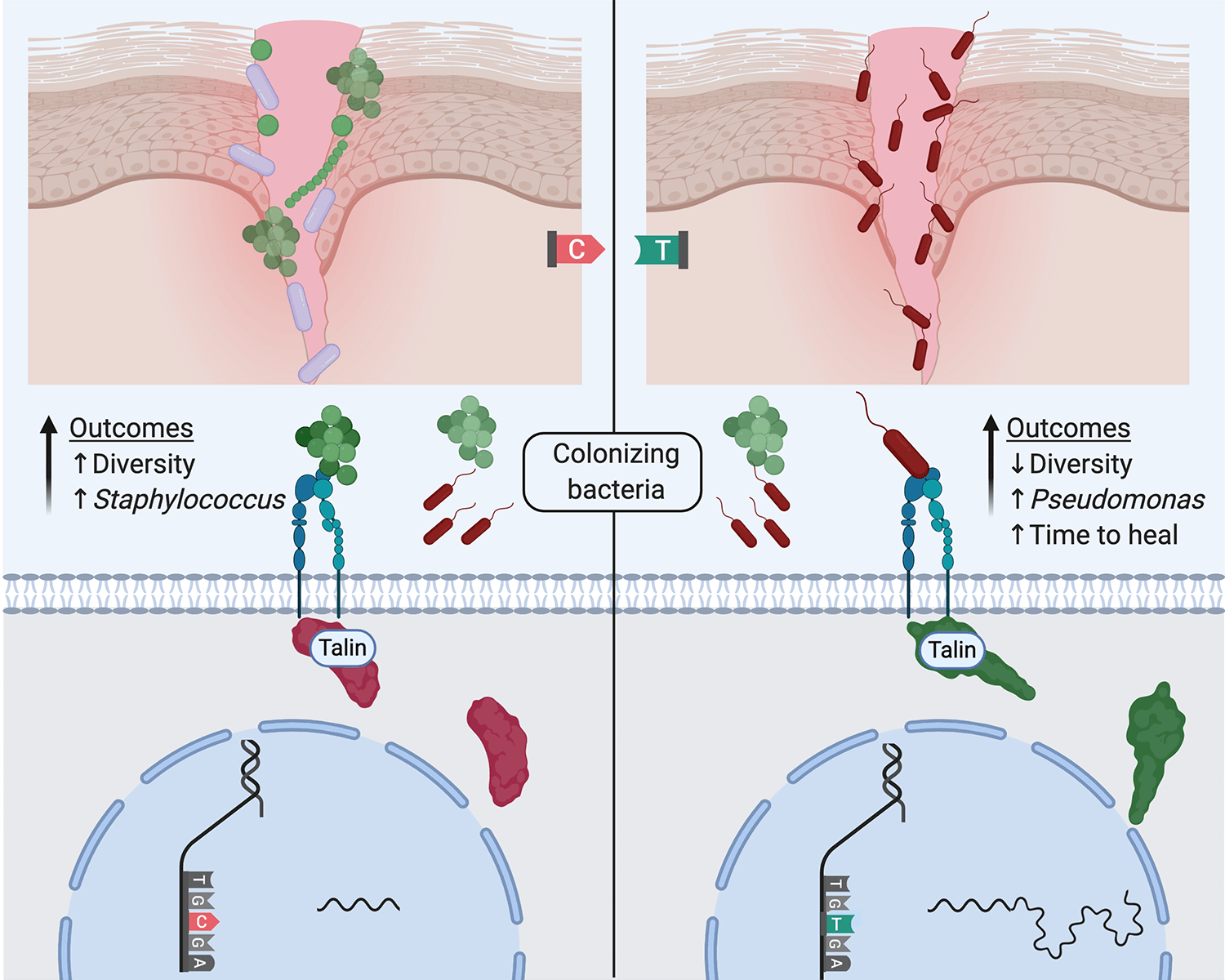
Tipton et al. provide initial insights into how patients' genetic variation shapes cellular expression phenotypes that associate with chronic wound microbiome diversity, species abundances and, in turn, healing. Here, a generalization of a working hypothesis emerging from the study is shown where a genetic difference influences alternative transcription of Talin2 leading to different focal adhesion dynamics that result in differential bacterial colonization of wounds.
Phillips said he looks forward to continuing his research at Texas Tech. His lab is developing a follow-up study that he hopes will collect enough information to create accurate predictive models. They also are working on a study exploring how a person's location in the U.S. shapes differences in chronic wound microbiomes.
"Texas Tech provides good support for research and is continually working for growth," Phillips said. "My research, like that of most others, has been generally enhanced by the academic freedom provided at the university. The Natural Science Research Laboratory is a premier Natural History Collection, and the samples archived at the Genetic Resources Collection have allowed me to design studies such as this one that would otherwise not have been possible. The hard work and creativity of doctoral student Craig Tipton were essential to the success of this project, as was collaboration with the laboratories of Nicole Phillips at UNTHSC and Kendra Rumbaugh at TTUHSC, Professor Todd Little in the Texas Tech College of Education, the SWRWCC and the NSRL."
The team of researchers included:
Department of Biological Sciences and the NSRL: Phillips and Tipton, who led the project from conception to experimental design and conducted data analysis, wet lab validation and structural equation modeling and wrote the final paper. The NSRL also assisted with curation and availability of samples and data curation.
SWRWCC: Wolcott, technical director Nicholas E. Sanford and information technology network administrator Clint Miller, who assisted with sample collection and data curation.
TTUHSC: Rumbaugh, a professor in the Department of Surgery and co-director of the Burn Center of Research Excellence; and post-doctoral researcher Derek Fleming, who conducted mouse model histological imaging.
Department of Microbiology, Immunology and Genetics at UNTHSC: Nicole Phillips, assistant
professor and director of the N. Phillips Lab; Gita Pathak and Talisa K. Silzer, who
were doctoral students during the project in the N. Phillips lab; and post-doctoral
researcher Jie Sun. The group assisted with patient genome fingerprinting.
Texas Tech Department of Educational Psychology and Leadership in the College of Education
and Optentia Research Focus Area at North-West University in Vanderbijlpark, South
Africa: Little, who assisted with structural equation modeling.
To read the complete study, visit the PLOS Pathogens website.
College of Arts & Sciences
-
Address
Texas Tech University, Box 41034, Lubbock, TX 79409-1034 -
Phone
806.742.3831 -
Email
arts-and-sciences@ttu.edu
