Dr. Huazhong Shi

Title: Professor
Education: Ph.D., Wuhan University, China, 1995
Research Associate, University of Arizona, 1999-2001, University of California, 2001-2003,
Purdue University, 2003-2004
Research Area: Biochemistry
Office: Chemistry 417
Phone: 806-834-7214
Email: huazhong.shi@ttu.edu
Webpage: Research Group
Interested in graduate study on molecular biology and biochemistry? Join the Shi lab! Applicants with strong Biology background are highly encouraged to apply. For more information, click here or contact Dr. Huazhong Shi.
Principal Research Interests
- Gene regulation in response to environmental stresses in plants
- Molecular mechanisms of plant salt tolerance
- Sulfonation of small molecules and plant stress response
Gene regulation in plant stress response
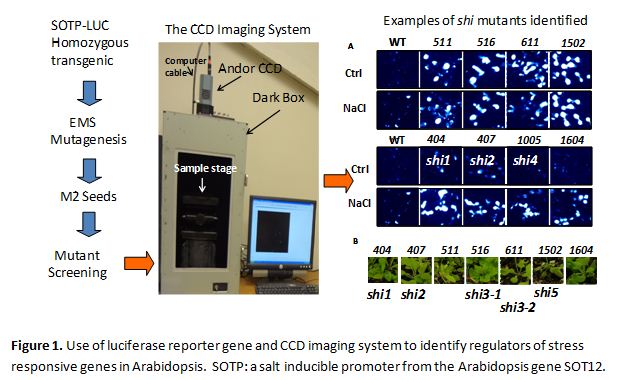
Fine regulation of gene expression is required for normal growth, development and adaptation to environmental stress conditions in plants. Many genes are repressed at normal growth conditions while activated by stresses. In order to identify regulator proteins mediating gene repression and activation, the Shi lab established a forward genetic screening for mutations affecting gene expression in response to stress conditions. The forward genetic approach utilized a stress-inducible promoter fused with the firefly luciferase reporter gene and a highly sensitive CCD camera to detect bioluminescence generated in small plant seedlings expressing the luciferase gene (Figure 1). By using this high throughput mutant screening system, the Shi lab has identified a number of mutants, designated shiny (shi in short) mutants, showing elevated expression of luciferase gene in response to abiotic stresses. Eight SHI genes have been cloned using map-based cloning. Two of the SHI genes encode proteins forming a complex to regulate stress-inducible gene expression through modulating RNA polymerase II CTD phosphorylation. Another three SHI proteins are mRNA splicing factors that are presumably components of repressor complex modulating stress-inducible gene transcription. Two of the eight SHI genes encode proteins functioning in vesicle trafficking. The Shi lab is employing all available means to study the functions of these SHI genes in order to gain deep understanding of stress-inducible gene repression and activation. In addition, the Shi lab initiated a forward genetic screening for mutations altering heat stress responsive gene expression. Positional cloning of the mutant genes is currently underway.
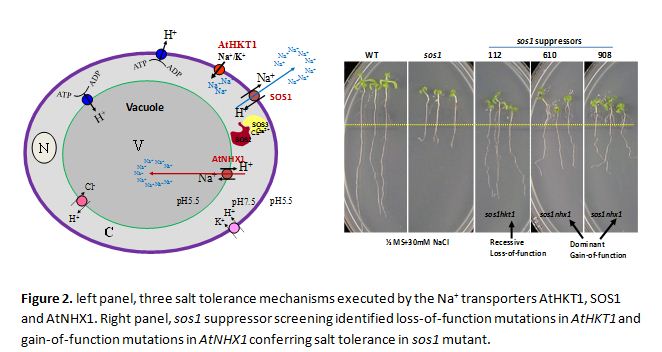
Na+ transport and plant salt tolerance
Plants possess three cellular mechanisms to reduce Na+ toxicity, i.e. restricted Na+ entry, Na+ exclusion, and Na+ compartmentation into the vacuole. These three cellular mechanisms are executed by three membrane transporters named AtHKT1, SOS1 and AtNHX1 in Arabidopsis (Figure 2). Through a genetic screening for suppressors of Na+ hypersensitive mutant sos1, the Shi lab identified loss-of-function mutations in AtHKT1 and gain-of-function mutations in AtNHX1 that can suppress the salt sensitivity of sos1 mutant. Single, double and triple mutants with mutations in these three important salt tolerance determinants have been created. The functions of these transporters and their coordination in Na+ uptake, long-distance transport, and redistribution in plants have been studied. Structure-function analysis of the dominant gain-of-function mutations in AtNHX1 and molecular design of superactive AtNHX1 transporters based on such analysis will be our future focus. The ultimate goal of this project is to create salt tolerant crops capable of growing in marginal lands with high salinity to maximize land usage and to secure world food supply.
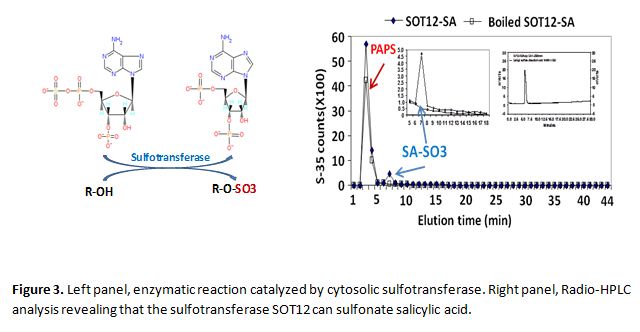
Sulfonation of small molecules and its function in plant stress response
Sulfonation of small molecules is an enzymatic process in living organisms catalyzed by sulfotransferases to transfer a sulfonate group from the universal donor 3'-phosphoadenosine 5'-phosphosulfate (PAPS) to the hydroxyl group of various molecules (Figure 3). Sulfonation changes the physiochemical properties and the biological activity of molecules, thus influencing the physiology of organisms. In human, sulfonation of small molecules plays important roles in detoxification of toxic compounds and modulation of steroid hormones. In Arabidopsis, 18 cytosolic sulfotransferases (SOTs) have been identified based on sequence similarity. The Shi lab carried out a genetic and biochemical analysis of the SOT12 and found that SOT12 can sulfonate the plant hormone salicylic acid (Figure 3) and xenobiotic compounds. The role of SOT12 in both biotic and abiotic stress response and tolerance is being studied. The Shi lab is also interested in the functions of other SOTs, in particular, their roles in stress response.
Representative Publications
- "NTR1 is involved in heat stress tolerance through mediating expression regulation and alternative splicing of heat stress genes in Arabidopsis"”, He L, Wu Q, Jin Y, Fan Y, Shi H, Wang Y, Yang W. Front Plant Sci, 2023, 13:1082511.
- "Coevolution of tandemly repeated hlips and RpaB-like transcriptional factor confers desiccation tolerance to subaerial Nostoc species"”, Xu HF, Dai GZ, Bai Y, Shang JL, Zheng B, Ye DM, Shi H, Kaplan A, Qiu BS. Proc Natl Acad Sci USA, 2022, 119: e2211244119.
- "SUMO E3 ligase SIZ1 negatively regulates arsenite resistance via depressing GSH biosynthesis in Arabidopsis"”, Hong Y, Chen Y, Shi H, Kong X, Yao J, Lei M, Zhu J-K, Wang Z. Stress Biology, 2022, 2: 9.
- "Acetylproteomics analyses reveal critical features of lysine-ε-acetylation in Arabidopsis and a role of 14-3-3 protein acetylation in alkaline response"”, Guo J, Chai X, Mei Y, Du J, Du H, Shi H, Zhu J-K, Zhang H. Stress Biology, 2022, 2: 1.
- "Gain-of-function mutations of AtNHX1 suppress sos1 salt sensitivity and improve salt tolerance in Arabidopsis"”, Pabuayon ICM, Jiang J, Qian H, Chung J-S, Shi H. Stress Biology, 2021, 1: 14
- "Comparative physiological and transcriptomic analysis reveals salinity tolerance mechanisms in Sorghum bicolor (L.) Moench"”, Ukwatta J, Pabuayon ICM, Park J, Chen J, Chai X, Zhang H, Zhu J-K, Xin Z, Shi H. Planta, 2021, 254: 98.
- "SWO1 modulates cell wall integrity under salt stress by interacting with importin ɑ in Arabidopsis"”, Wang Z, Wang M, Yang C, Zhao L, Qin G, Peng L, Zheng Q, Nie W, Song C-P, Shi H, Zhu J-K, Zhao C. Stress Biology, 2021, 1: 9.
- "HISTONE DEACETYLASE 6 suppresses salicylic acid biosynthesis to repress autoimmunity"”, Wu Z, He L, Jin Y, Chen J, Shi H, Wang Y, Yang W. Plant Physiol, 2021, 187: 2592-2607.
- "The Arabidopsis spliceosomal protein SmEb modulates ABA responses by maintaining proper alternative splicing of HAB1"”, Hong Y, Yao J, Shi H, Chen Y, Zhu JK, Wang Z. Stress Biology, 2021, 1:4.
- "Initiation and amplification of SnRK2 activation in abscisic acid signaling", Lin Z, Li Y, Wang Y, Liu X, Ma L, Zhang Z, Mu C, Zhang Y, Peng L, Xie S, Song C-P, Shi H, Zhu J-K, Wang P. Nat Commun, 2021, 12: 2456.
- "COP1 Promotes ABA-induced stomatal closure by modulating the abundance of ABI/HAB and AHG3 phosphatases", Chen Q, Bai L, Wang W, Shi H, Botella JR, Zhan Q, Liu K, Yang H-Q, Song C-P. New Phytol, 2021, 229: 2035-2049.
- "Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication", Wang Z, Hong Y, Li Y, Shi H, Yao J, Liu X, Wang F, Huang S, Zhu G, Zhu J-K. Plant Biotechnol J, 2021, 19: 20-22.
- "Selenium supply alters the subcellular distribution and chemical forms of cadmium and the expression of transporter genes involved in cadmium uptake and translocation in winter wheat (Triticum aestivum)", Zhu J, Zhao P, Nie Z, Shi H, Li C, Wang Y, Qin S, Qin X, Liu H. BMC Plant Biol, 2020, 20: 550.
- "Reciprocal regulation between nicotinamide adenine dinucleotide metabolism and abscisic acid and stress response pathways in Arabidopsis", Hong Y, Wang Z, Shi H, Yao J, Liu X, Wang F, Zeng L, Xie Z, J-K Zhu. PLoS Genet, 2020, 16: e1008892.
- "TPST is involved in fructose regulation of primary root growth in Arabidopsis thaliana", Zhong Y, Xie J, Wen S, Wu W, Tan L, Lei M, Shi H, Zhu J-K. Plant Mol Biol, 2020, 103: 511-525.
- "Plant abiotic stress response and nutrient use efficiency", Gong Z, Xiong L, Shi H, Yang S, Herrera-Estrella LR, Xu G, Chao DY, Li J, Wang PY, Qin F, Li J, Ding Y, Shi Y, Wang Y, Yang Y, Guo Y, Zhu J-K. Sci China Life Sci, 2020, 63: 635-674.
- "Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter", Wang Z, Hong Y, Zhu G, Li Y, Niu Q, Yao J, Hua K, Bai J, Zhu Y, Shi H, Huang S, Zhu J-K. EMBO J, 2020, 39: e103256.
- "Dehydration-induced DnaK2 chaperone is involved in PSII repair of a desiccation-tolerant Cyanobacterium", Xu H-F, Dai G-Z, Ye D-M, Shang J-L, Song W-Y, Shi H, and Qiu B-S. Plant Physiol, 2020, 182: 1991-2005.
- "The plasma‐membrane polyamine transporter PUT3 is regulated by the Na+/H+ antiporter SOS1 and protein kinase SOS2", Chai H, Guo J, Zhong Y, Hsu C-C, Zou C, Wang P, Zhu J-K, Shi H. New Phytol, 2020, 226: 785-797.
- "Comparative transcriptomics of stem bark reveals genes associated with bast fiber development in Boehmeria nivea L. gaud (ramie)", Xie J, Li J, Jie Y, Xie D, Yang D, Shi H, Zhong Y. BMC Genomics, 2020, 21: 40.
- "STCH4/REIL2 confers cold stress tolerance in Arabidopsis by promoting rRNA processing and CBF protein translation", Yu H, Kong X, Huang H, Wu W, Park J, Yum D-J, Lee B-H, Shi H, Zhu J-K. Cell Reports, 2020, 30: 229–242.
- "Pst DC3000 infection alleviates subsequent freezing and heat injury to host plants via a salicylic acid-dependent pathway in Arabidopsis", Tuang ZK, Wu Z, Jin Y, Wang Y, Phyo Zin Oo P, Zuo G, Shi H, Yang W. Plant Cell Environ, 2020, 43: 801-817.
- "Two chloroplast proteins negatively regulate plant drought resistance through separate pathways", Hong Y, Wang Z, Liu X, Yao J, Kong X, Shi H, Zhu J-K. Plant Physiol, 2020, 182: 1007-1021
- "The DEAD-box RNA Helicase SHI2 Functions in repression of salt-inducible genes and regulation of cold-inducible gene splicing", Wang B, Chai H, Zhong Y, Shen Y, Yang W, Chen J, Xin Z, Shi H. J Exp Bot, 2020, 71: 1598-1613.
- "A nematode sterol C4α-methyltransferase catalyzes a new methylation reaction responsible for sterol diversity", Zhou W, Fisher PM, Vanderloop BH, Shen Y, Shi H, Maldonado AJ, Leaver DJ, Nes WD. J Lipid Res, 2020, 61:192-204
- "Nitrogen supply enhances zinc uptake and root-to-shoot translocation via up-regulating the expression of TaZIP3 and TaZIP7 in winter wheat (Triticum aestivum)", Nie Z, Zhao P, Shi H, Wang Y, Qin S, Liu H. Plant Soil, 2019, 444: 501–517.
- "The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice", Miao C, Wang Z, Zhang L, Yao J, Hua K, Liu X, Shi H, Zhu J-K. Nat Commun, 2019, 10: 3822.
- "Cold stress activates disease resistance in Arabidopsis thaliana through a salicylic acid dependent pathway", Wu Z, Han S, Zhou H, Tuang ZK, Wang Y, Jin Y, Shi H, Yang W. Plant Cell Environ, 2019, 42: 2645-2663.
- "The Flowering Repressor SVP Confers Drought Resistance in Arabidopsis by Regulating Abscisic Acid Catabolism", Wang Z, Wang F, Hong Y, Yao J, Ren Z, Shi H, Zhu J-K. Mol Plant, 2018, 11: 1184-1197.
- "Knock-down of rice microRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development", Zhang J, Zhang H, Srivastava AK, Pan Y, Bai J, Fang J, Shi H, Zhu J-K. Plant Physiol, 2018, 176: 2082-2094.
- "Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1", Ren Z, Wang Z, Zhou XE, Shi H, Hong Y, Cao M, Chan Z, Liu X, Xu HE, Zhu J-K. Sci Rep, 2017, 7: 14022.
- "Histone deacetylase 6 represses pathogen defense responses in Arabidopsis thaliana", Wang Y, Hu Q, Wu Z, Wang H, Han S, Jin Y, Zhou J, Zhang Z, Jiang J, Shen Y, Shi H, Yang W. Plant Cell Environ, 2017, 40: 2972–2986.
- "Salt tolerance response revealed by RNA-Seq in a diploid halophytic wild relative of sweet potato", Luo Y, Reid R, Freese D, Li C, Watkins J, Shi H, Zhang H, Loraine A, Song B-H. 2017, Sci Rep, 7: 9624.
- "Polyamine and paraquat transport assays in Arabidopsis seedling and callus", Chai H, Shen Y, Shi H. Bio-protocol, 2017, 7(15).
- "Cellular polyamines modulate mRNA stability", Chai H, Yang W, Shi H. Plant Signal Behav, 2017, 12:10, e1323163.
- "Overexpression of PP2A-C5 that encodes the catalytic subunit 5 of protein phosphatase 2A in Arabidopsis confers better root and shoot development under salt conditions", Hu R, Zhu Y, Wei J, Chen J, Shi H, Shen G, Zhang H. Plant Cell Environ, 2017, 40: 150-164.
- "Improved salt tolerance of medicinal plant Codonopsis pilosula by Bacillus amyloliquefaciens GB03", Han Q-Q, Wu Y-N, Gao H-J, Xu R, Pare PW, Shi H, Zhao Q, Li H-R, Khan SA, Wang Y-Q, Suo-Min Wang S-M, Zhang J-L. Acta Physiol Plant, 2017, 39:35.
- "Soybean Na+/H+ antiporter GmsSOS1 enhances antioxidant enzyme activity and reduces Na+ accumulation in Arabidopsis and yeast cells under salt stress", Zhao X, Wei P, Liu Z, Yu B, Shi H. Acta Physiol Plant, 2017, 39:19.
- "The Nuclear Encoded Chloroplast Protein HCF106 and THF1 Negatively Regulate Drought Resistance in Arabidopsis", Wang Z, Wang F, Hong Y, Huang J, Shi H, Zhu J-K. Plant Physiol, 2016, 172: 2491-2503.
- "The Arabidopsis polyamine transporter LHR1/PUT3 modulates heat responsive gene expression by enhancing mRNA stability", Shen Y, Ruan Q, Chai H, Yuan Y, Yang W, Chen J, Xin Z, Shi H. Plant J, 2016, 88: 1006-1021.
- “De Novo Assembly and Characterization of Gleditsia sinensis Transcriptome, Genes Identification and SSR Mining”, Han S, Wu Z, Wang X, Huang K, Jin Y, Yang W, Shi H. Genet Mol Res, 2016, 15 (1): gmr.15017740,DOI http://dx.doi.org/10.4238/gmr.15017740.
- “Induced growth promotion and higher salt tolerance in the halophyte grass Puccinellia tenuiflora by beneficial rhizobacteria”, Niu S-Q, Li H-R, Pare PW, Aziz M, Wang S-M, Shi H, Li J, Han Q-Q, Guo S-Q, Li J, Guo Q, Ma Q, Zhang J-L. Plant & Soil, 2016, 407:217–230.
- “Research advances in higher plant adaptation to salt stress”, Zhang J-L, Li H-R, Guo S-Y, Wang S-M, Shi H, Han Q-Q, Bao A-K, Ma Q. Acta Prataculturae Sinica, 2015, 24: 220-236.
- “The role of promoter cis-element, mRNA capping, and ROS in the repression and salt-inducible expression of AtSOT12 in Arabidopsis”. Chen J, Wang B, Chung J-S, Chai H, Liu C, Ruan Y, Shi H. Front Plant Sci, 2015, 6:974. doi:10.3389/fpls.2015.00974.
- "RNA-Seq Analysis for Transcriptome Assembly, Gene Identification and SSR Mining in Ginkgo (Ginkgo biloba L.)". Han S, Wu Z, Jin Y, Yang W, Shi H. Tree Genet Genomes, 2015, 11:37, doi: 10.1007/s11295-015-0868-8.
- "Detoxification function of the Arabidopsis sulfotransferase AtSOT12 by sulfonation of xenobiotics", Chen J, Gao L, Baek DW, Liu C, Ruan Y, Shi H. Plant Cell Environ, 2015, 38: 1673-1682, doi: 10.1111/pce.12525.
- "Bacillus crassostreae sp. nov., isolated from an oyster (Crassostrea hongkongensis)", Chen J, Tian X, Ruan Y, Yang Y, He Z, Tang S, Li W, Shi H, Chen Y. Int J Syst Evol Microbiol, 2015, 65: 1561-1566, doi:10.1099/ijs.0.000139.
- "GmFLD, a soybean homolog of the autonomous pathway gene FLOWERING LOCUS D, promotes flowering in Arabidopsis thaliana", Hu Q, Jin Y, Shi H, Yang W. BMC Plant Biol, 2014, 14:263.
- "The Arabidopsis RNA Binding Protein with K Homology Motifs, SHINY1, Interacts with the C-terminal Domain Phosphatase-like 1 (CPL1) to Repress Stress-Inducible Gene Expression", Jiang, J.; Wang, B.; Shen, Y.; Wang, H.; Feng, Q.; Shi, H. PLoS Genet, 2013, 9(7): e1003625. doi:10.1371/journal.pgen.1003625.
- "Physiological and molecular mechanisms of plant salt tolerance", Zhang, J.; Shi, H. Photosynth Res, 2013, 115:1-22.
- “A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to derepression of seed-specific gene expression” Veerappan, V; Wang, J; Kang, M; Lee, J; Tang, Y; Jha, AK; Shi, H; Palanivelu, R; Allen, RDPlanta2012, 236,1-17.
- “Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance” Baek, DW; Jiang, J; Chung, JS; Wang, B; Chen, J; Xin, Z; Shi, H.Plant Cell Physiol.2011, 52, 149-161.
- “A Stress-inducible Sulfotransferase Sulfonates Salicylic Acid and Confers Pathogen Resistance in Arabidopsis” Baek, DW; Pathange, P; Chung, JS; Jiang, J; Gao, L; Oikawa, A; Hirai, MY; Saito, K; Pare, PW; Shi, H. Plant Cell Environ.2010, 33,1383-1392.
- “HOS3, an ELO-like gene, inhibits effects of ABA and implicates a S-1-P/ceramide control system for abiotic stress response in Arabidopsis thaliana” Quist, TM; Sokolchik, I; Shi, H; Joly, RJ; Bressan, RA; Maggio, A; Narsimhan, M; Li, X. Mol. Plant2009, 2,138-151.
- "Signaling control of SOS1 mRNA stability". Jiang, J; Shi, H. Plant Signaling and Behavior2008, 3, 687-688.
- "Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1". Zhang, H; Kim, M-S; Sun, Y; Dowd, SE; Shi, H; Pare, PW. Molecular Plant-Microbe Interaction2008, 21, 737-744.
- "Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance". Zhu, J; Jeong, JC; Zhu, Y; Sokolchik, I; Miyazaki, S; Zhu, J-K; Hasegawa, PM; Bohnert, HJ; Shi, H; Yun, D-J; Bressan, RA. Proc Natl Acad Sci USA2008, 105, 4945-4950
- "Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis". Chung, JS; Zhu, JK; Bressan, RA; Hasegawa, PM; Shi, H. Plant J.2008,53, 554-565.
- "Integration of Ca2+ in plant drought and salt stress signal transduction pathways". Shi, H. In Advances in molecular breeding towards salinity and drought tolerant (eds. Jenks MA, Hasegawa PM, Jain SM).Springer 2007, pp. 141-182.
- "Isolation and characterization of shs1, a sugar-hypersensitive and ABA-insensitive mutant with multiple stress responses". Inan, G; Goto, F; Jin, JB; Rosado, A; Koiwa, H; Shi, H; Hasegawa, PM; Bressan, RA; Maggio, A; Li, X. Plant Mol Biol2007, 65, 295-309.
- "An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response". Zhu, J; Fu, X; Koo, YD; Zhu, JK; Jenney, FE Jr; Adams, MW; Zhu, Y; Shi, H; Yun, DJ; Hasegawa, PM; Bressan, RA. Mol Cell Biol2007,27, 5214-5224.
- "Salt stress affects cortical microtubule organization and helical growth in Arabidopsis". Shoji, T; Suzuki, K; Abe, T; Kaneko, Y; Shi, H; Zhu, JK; Rus, A; Hasegawa, PM; Hashimoto, T. Plant Cell Physiol.2006, 47, 1158-1168.
- "RNA Extraction". Shi, H; Bressan, R. In Methods in Molecular Biology, Vol. 323, Arabidopsis Protocols (2nd ed) (Eds Salinas J, Sanchez-Serrano J), Humana Press, New Jersey, 2006, pp 345-348.
- "Sodium", Shi, H.; Bressan, R.; Hasegawa, P.M.; Zhu, J.-K. In Plant Nutritional Genomics (Broadley. M; White, P., Eds), Blackwell Publishing, London 2004.
- "An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway", Zhu, J.; Shi, H.; Lee, B.H.; Damsz, B.; Cheng, S.; Stirm, V.; Zhu, J.K.; Hasegawa, P.M.; Bressan, R.A. Proc Natl Acad Sci USA 2004, 101, 9873-9878.
- "Topological analysis of a plant vacuolar Na/H antiporter reveals a luminal C-terminus that regulates the antiporter activity", Yamaguchi, T.; Apse, M.P.; Shi, H.; Blumwald, E. Proc Natl Acad Sci USA 2003, 100, 12510-12515.
- "The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion", Shi, H.; Kim, Y.-S.; Guo, Y.; Zhu, J.-K. Plant Cell 2003, 15, 19-32.
- "Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis", Shi, H.; Lee, B.; Wu, S.-J.; Zhu, J.-K. Nat Biotechnol. 2003, 21, 81-85.
- "Regulation of the vacuolar Na+/H+ antiporter gene AtNHX1 expression by salt stress and abscisic acid", Shi, H.; Zhu, J.-K. Plant Mol Biol. 2002, 50, 543-550.
- "Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis", Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.K.; Pardo, J.M. Proc Natl Acad Sci USA 2002, 99, 9061-9066.
- "SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis", Shi, H.; Zhu, J.-K. Plant Physiol. 2002, 129, 585-593.
- "The Arabidopsis salt overly sensitive 4 mutants uncover a critical role for vitamin B6 in plant salt tolerance", Xiong, L.; Stevenson, B.; Lu, T.; Zhu, J.-K. Plant Cell 2002, 14, 575-588.
- "The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants", Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. Plant Cell 2002, 14, 465-477.
- "The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter", Shi, H.; Ishitani, M.; Kim, C.; Zhu, J. –K. Proc Natl Acad Sci USA 2000, 97, 6896-6901.
Department of Chemistry & Biochemistry
-
Address
1204 Boston Avenue, Lubbock, TX 79409-1061 -
Phone
806.742.3067
